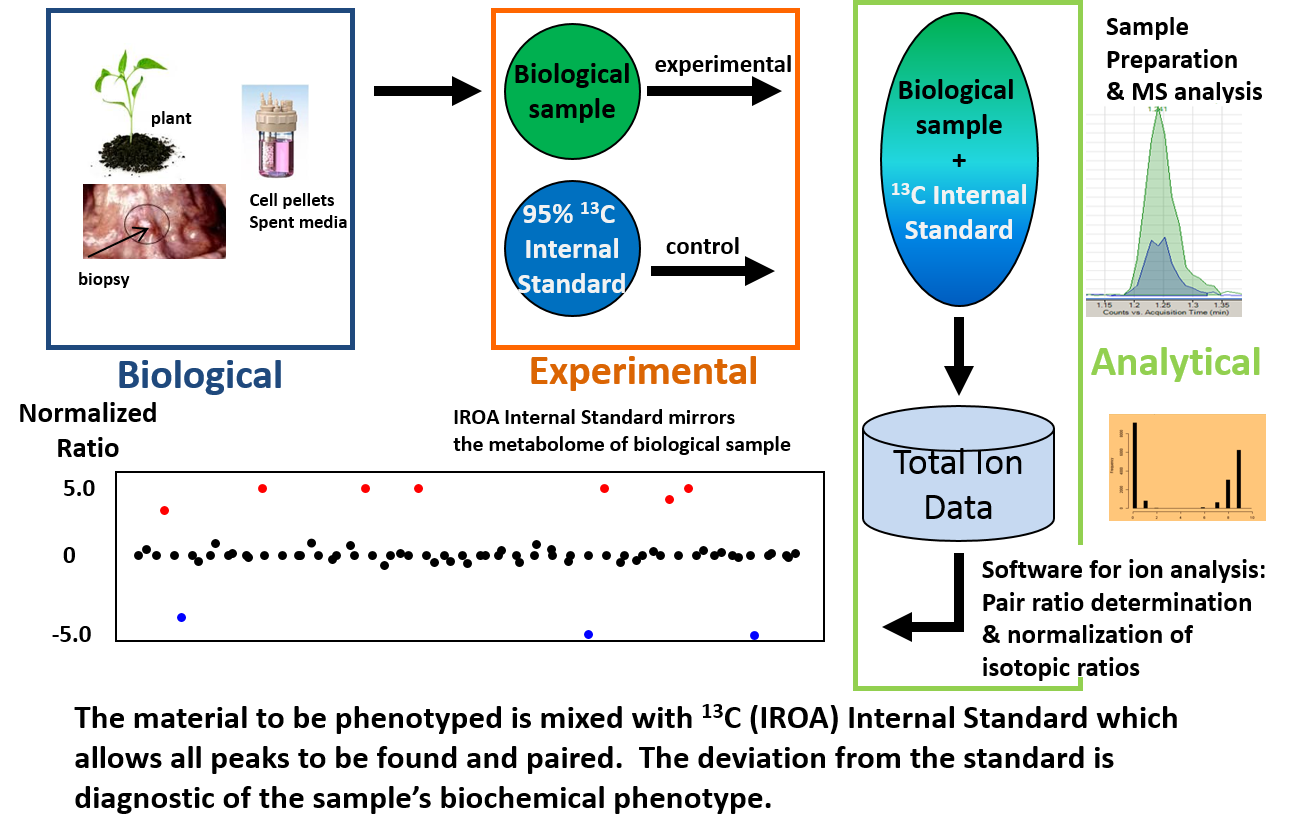
In the Basic IROA experiment, both Control and Experimental samples are labeled in-vitro. There are many cases where it is not practical or possible to label the Experimental sample, for example, when working with tissue samples, performing large fermentation runs, or propagating field-grown plants. In these instances, the IROA “Phenotypic” experiment may be applied. In the Phenotypic Protocol the Control or complex “Internal Standard” is isotopically labeled with IROA U-13C media and used to spike into Experimental samples.
An ideal Standard is one that represents the entire metabolome of the fluid or tissue under study. For example, if interrogating the metabolome in pancreatic tissue, quantification of metabolites can be achieved using an IROA-grown/labeled pancreatic cell line as a complex Internal Standard.
The IROA Phenotypic Experiment. The material to be phenotyped i.e. biopsy tissue, plant material, cells, etc. is mixed with the labeled 13C IROA- Internal Standard. The IROA ClusterFinder software will find all IROA-labeled compounds and use these peaks to locate and identify the associated paired natural abundance peaks in the Experimental samples. This allows for a complex targeted analysis.
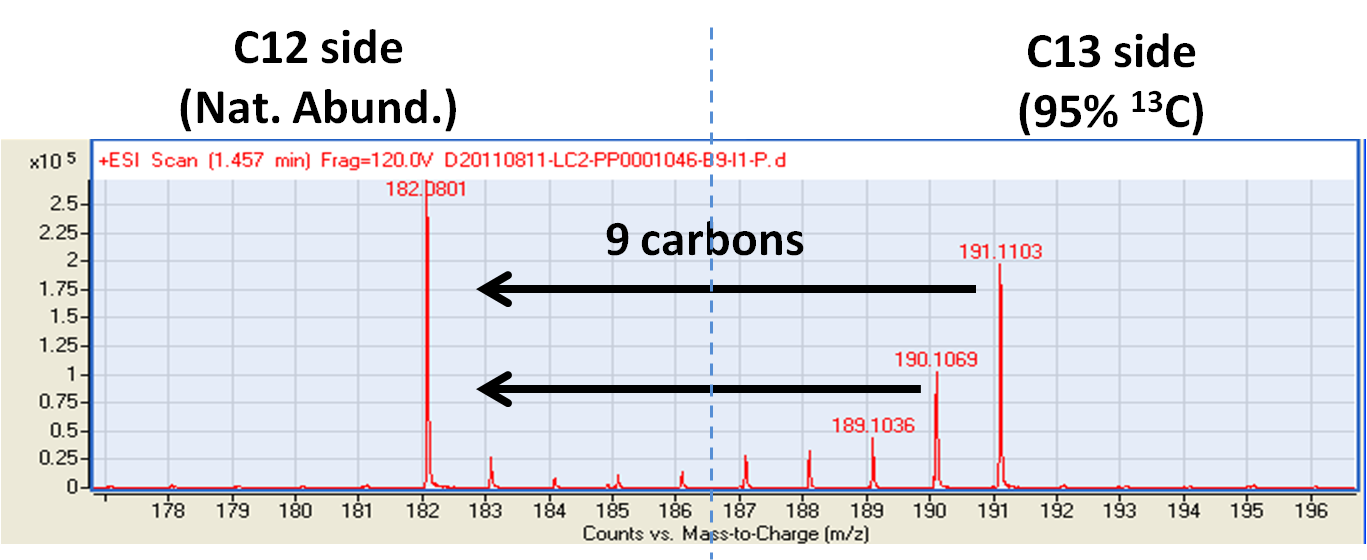
Using an IROA-labeled Standard, all of peaks may be easily identified according to presence of their characteristic M-1 peak. The natural abundance peaks corresponding to each Standard peak may be readily identified because even though they do not carry any isotopic labeling, their exact mass and position are established relative to the Standard.
If the pooled Standard is well characterized and the compounds that are present in it have already been identified, then it will be able to be used as a point of comparison for all of its contained compounds. Artifacts will have no match in the Standard and are eliminated by ClusterFinder in a final dataset. In a typical experimental IROA dataset the ratio of the peak areas represent the relative deviation of the metabolic pool sizes brought about by the experimental condition. In a Phenotypic experiment, all experimental samples are measured relative to the Standard; therefore the phenotype is defined by the deviation from the Standard
View the Application Note: Differential Metabolomic Profiling of Maize Genotypes under Drought-Stressed Conditions using IROA® (Isotopic Ratio Outlier Analysis)
A tutorial on the Phenotypic Experiment can be viewed here.

