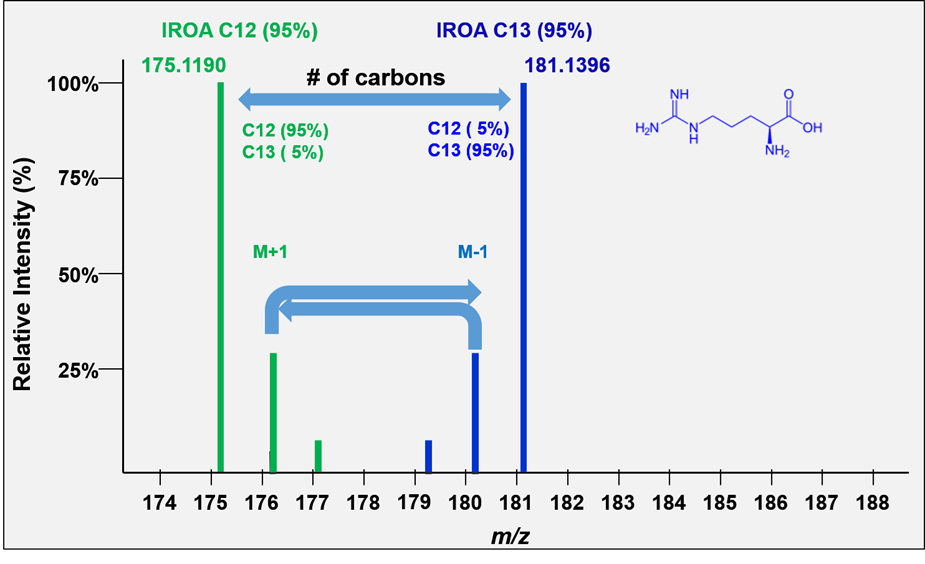
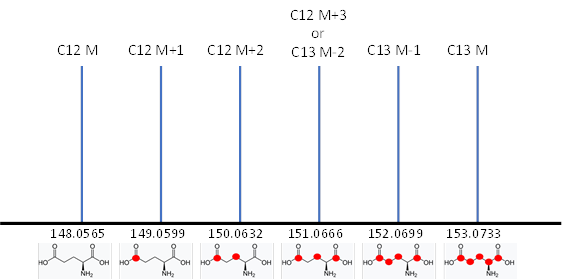
All molecules with the same number of carbons will show the same pattern of peaks but will differ in the mass of their monoisotopic peaks according to the remainder of the formula. We refer to these peak height patterns as the peak “isotopic envelopes”. These envelopes are diagnostic for each formula.
The IROA carbon envelope shapes are readily and exactly calculable. The defining feature of the IROA carbon envelope is the mass of both monoisotopic peaks and the mass difference between them. The mass difference between the monoisotopic peaks is always a multiple of the mass of a neutron (~1.00335 amu). The additional peaks discussed above contribute to the extended isotopic envelope (M+2, M+3 etc., M-2, M-3 etc., see Figure 4) and the IROA ClusterFinder software can easily identify these peaks by their mass difference (the mass difference between a 13C and 12C isotope).
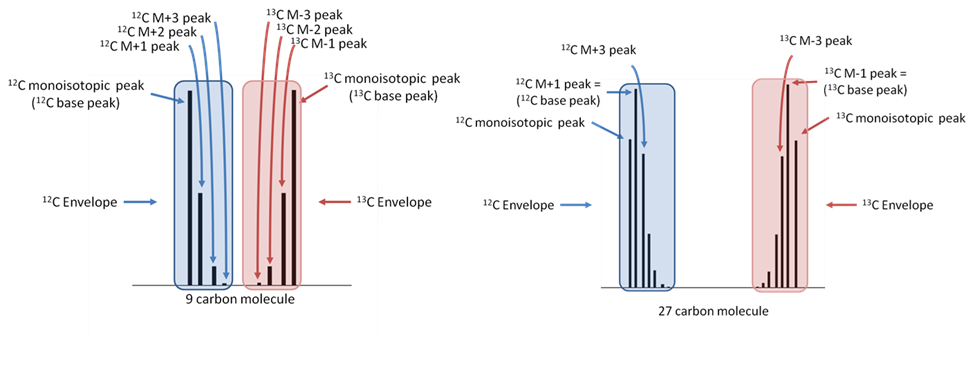
Figure 4. IROA isotopic envelopes (clusters) illustrating parts for a 9 carbon and 27 carbon molecule labeled with 5%13C:95%13C. Note the base peak shift. The monoisotopic peak for the 27-carbon molecule (right side of figure) is not the most abundant peak in the envelope.
- Isomers, isotopomers, and isotopolog patterns become complex very quickly. No two isotopologs are the same, and any isotopolog will generally contain more than one isotopomer and each contains different collections of isotopomers. We find this exact language to be useful however cumbersome.
- For IROA, we generally use two different isotopic distributions creating patterns that represent different isotopic balances, for example 1.1%13C:95%13C or 5%13C:95%13C. One of these, usually based on the C13 monoisotopic peak, will be the internal standard.
- For each formula all these isotopic patterns must use the same isotopolog ladders, but they will fill them differently.
- We believe it is easier to combine the embodiments of the isotopomers and isotopologues under the rubric of an “isotopic envelope”.
- There are two isotopic envelopes in any IROA sample.
Carbon is the only element that has an exact unit mass. This is because carbon is defined to be exactly 12.000000 amu. One outcome of this is that the exact unit masses of all other elements have slightly different “defects”, i.e., the fractional number beyond the decimal point. The mass “defect” is the difference between nominal mass (mass of the most abundant elemental isotope; for a molecule, the sum of the nominal masses of the constituent elements; i.e. H20=18) and the monoisotopic mass (exact mass) of an atom or molecule. These defects can only be added together a certain number of ways to get a specific mass as seen by a mass spectrometer.
Figure 5 shows an example of using mass defect to determine molecular formula. Using IROA we can calculate residual mass of all the other elements using both C12 and C13 base peaks. Since both the C12 and C13 base peaks share the same formula, the error on this residual value can be minimized by averaging the two residual masses.
With reasonably high-resolution mass MS (mass accuracy of 50 ppm or grater) and the knowledge of the number of carbons in a molecule (the distance between the C12 and C13 monoisotopic peaks), generally only one formula exists for molecules with masses below 500 amu. In the few cases where more than a single formula is possible, the use of mass defect residual tables can often resolve any ambiguity.
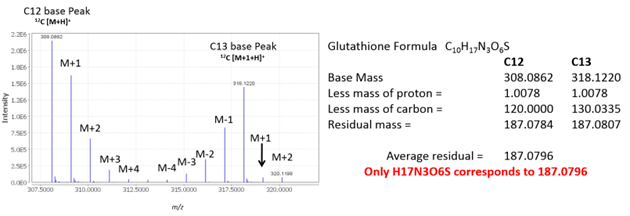
Figure 5. Calculation of average residual mass (mass defect) for glutathione.
Fundamental to the IROA concepts (and inherent in the name Isotopic Ratio Outlier Analysis) is the fact that the ratio of the C-12 envelope to the C-13 envelope is unaffected by suppression even though both the C-12 and C-13 isotopomeric sets may be strongly suppressed. This has afforded a mechanism for suppression correction that has been built into ClusterFinder. Once suppression is corrected, a Dual MSTUS[1] algorithm is employed to provide a very accurate mechanism for the normalization of samples against sample-to-sample variances. This version of ClusterFinder outputs three values: 1) the raw (suppressed) values observed; 2) a suppression-corrected value; and 3) a normalized (suppression-corrected and normalized) value.
Summary
- All isotopomers are isotopic isomers that share a single mass. There is no positional constraint.
- All isotopologs contain different numbers of isotopes but otherwise share a formula.
- In IROA there are always contributions from two different sources, usually an analyte and an internal standard. The peaks from both sources will superimpose onto the same ladder.
- An isotopolog ladder exists between the two monoisotopic peaks (from 2 different sources), and this ladder is diagnostic for the molecular formula that it is derived from.
- The collection of peaks donated by each source is grouped into a collection of isotopologs that is most easily identified as an isotopic envelope.
- Accurate quantitation requires summing all of the peaks in each envelope.
[1] Warrack BM, Hnatyshyn S, Ott KH, Reily MD, Sanders M, et al. (2009) “Normalization strategies for metabonomic analysis of urine samples.”, J Chromatogr B Analyt Technol Biomed Life Sci 877: 547–552.