OUR PRODUCTS
Products that make metabolomics easier
WORKFLOW KIT
MASS SPECTROMETRY
REMOVE SUPPRESSION AND NORMALIZE SAMPLES
OUR TECHNOLOGY
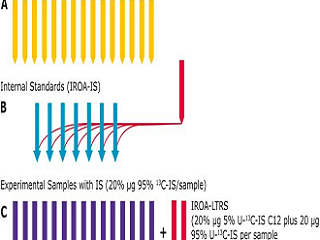
Learn how we use internal standards and companion algorithms as metabolic profiling tools to eliminate noise, analytical variability, and ion suppression. These tools lay the foundation to eliminate ion suppression and achieve metabolite normalization, two of the largest sources of error in metabolomics, making it possible to obtain clean, accurate, and reproducible datasets. Better data translates into better results. Better results mean more effective and countable outcomes. Our objective, at IROAtech, is to simplify clinical metabolomic measurements and streamline results for successful outcomes.
C13-labelled Yeast Semi-targeted Metabolomics Workflow

How IROA Works

IROA MLSDiscovery Software

Metabolomics Compound Identification
PUBLICATIONS
Read our publications on metabolic profiling technologies and learn about innovative and advanced solutions for metabolomics. Find out about the transformations that have automated the measurement of biochemicals in biological systems.
Metabolomics Research

ABOUT US
Metabolomics Company founded in 2010, in Ann Arbor, Michigan.
We are more than a metabolomics company, we are innovators simplifying metabolomics. We use stable isotope-labeled biochemical standards, software, and data analytics that correct ionization inefficiencies and instrumentation variances. We promote a streamlined metabolomics workflow employing advanced data analysis to make mass spectrally based measurements more accurate and reproducible so that results are consistent not only day-to-day but from batch-to-batch and across laboratories. From characterizing phenotypes to identifying biomarkers, we simplify clinical metabolomics to bring effective and meaningful results that you can rely on.
Listen To Webinar
The IROA TruQuant IQQ Cannabis metabolic profiling workflow provides a software solution together with a carefully formulated Cannabis Internal Standard for accurate simultaneous measurement of 100s of compounds.
Some of our Customers
Metric

NC State University

Senda

Stanford Health Care School of Medicine